Pangenome
Users have the possibility to explore pangenome datasets generated on 15 Musaceae accessions using Panache.
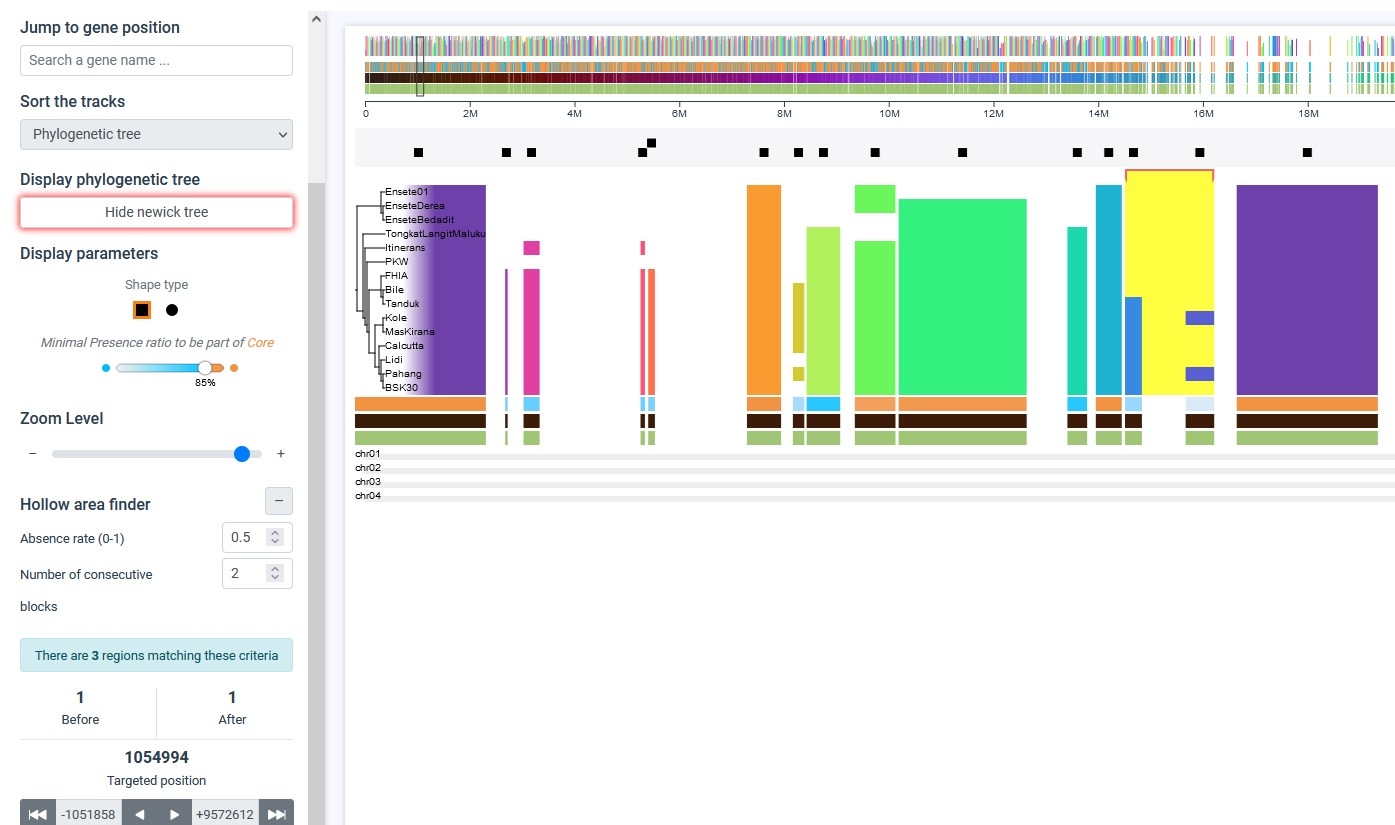
There is a range of options to explore presence absence variation (PAV) of the matrix. In the example below, we decided to use the hollow area finder to find at least 2 consecutive blocks (corresponding to genes in this dataset) that are absent to at least 50% of the sampling (highlighted in yellow). Then we sorted the individuals based on the phylogenetic tree. We can see that one block is consistently found on a clade, while the second bloc in only present in 2 individuals of the clade.
For more information about additional features, please look at
- Project website https://github.com/SouthGreenPlatform/panache
- Functionalities tour https://github.com/SouthGreenPlatform/panache/wiki/Functionalities-tour
- Publication https://doi.org/10.1093/bioinformatics/btab688




